1. How to use Jupyter Lab functionality¶
Jupyter Lab is a web application that allows you to use Python interactively through a browser, a Linux console, simple file manipulation, and graphical output of Python programs.
TSUBAME3 allows you to use Jupyter Lab directly through the TSUBAME portal, giving you a GPU-powered compute node that feels like Jupyter Lab on your laptop.
1.1. Start Jupyter Lab¶
You can create a dedicated job on the TSUBAME portal to launch Jupyter Lab.
For details, please refer to the TSUBAME Portal User's Guide.
Tips
You can display a pre-launched Jupyter Lab by selecting the [View] button of the corresponding job on the Web service use screen of TSUBAME portal.
1.2. Quit Jupyter Lab¶
Please select "Shut Down" from the File menu in Jupyter Lab.
If you select Logout, the job will remain running and you will continue to be charged.
You can also exit from the TSUBAME Portal's Web Service User Screen.
1.3. Data workspace¶
The only directory visible to Jupyter Lab will be the t3workspace directory in the user's home directory.
Also, the size of files that can be uploaded via the Jupyter web interface is limited, so if you want to upload large data files, use commands such as wget from the console, or consider using sftp, rsync, etc.
1.4. Install modules from PyPI¶
The Python environment in the Jupyter Lab is maintained separately from the command-line Python environment.
Modules can be installed and updated from the console in the Jupyter Lab with the following commands
python3 -m pip install --user module name
To install in the user directory, the --user is required.
-U option must be added when upgrading.
The above command can also be executed on the Notebook in a cell with the %%bash on the first line.
%%bash
python3 -m pip install --user module name
Warning
After the module is installed, the kernel (Python environment) needs to be restarted.
Select Restart Kernel from the Kernel menu (or type 0 twice).
1.5. Create custom kernel¶
When you install some Python modules that use GPUs, you need to define a kernel (Python environment) that is loaded with software such as CUDA.
This section shows how to create a custom kernel with CuPy using CUDA, CuDNN, and NCCL as an example.
Info
There is a Notebook(in Japanese) with descriptions and commands for this section that you can use.
t3jpttools module is provided in the Jupyter Lab of TSUBAME3, and the following functions are available.
- create_kernel(kernel name, list of modules): to define a new custom kernel
- list_kernel(): Displays a list of kernels
- delete_kernel(kernel name): delete a custom kernel
For example, to create a custom kernel that uses CUDA 10.2.89, CuDNN 7.6, or NCCL 2.4.2, run the following command in the Notebook cell.
from t3jpttools import create_kernel
create_kernel('python3-cuda','cuda/10.2.89 cudnn/7.6 nccl/2.4.2')
After executing create_kernel() and waiting for a few tens of seconds, you will be able to select the created kernel from the top right of the screen.
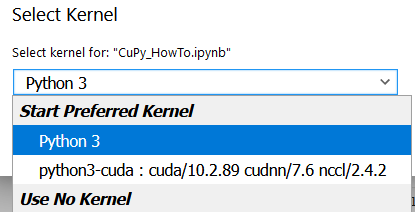
By switching the kernel, you will be able to install and execute modules that depend on CUDA, such as CuPy.
Info
To change the Python version used in Notebook, specify jupyterlab/2.1.0-py383 when executing create_kernel().
This is a Jupyter module that includes Python 3.8.3, please check the existence with module avail command beforehand.
Example: create_kernel('test','jupyterlab/2.1.0-py383 cuda/10.2.89 cudnn/7.6 nccl/2.4.2')